-
Notifications
You must be signed in to change notification settings - Fork 20
Centromere Telomere locations
BRASS needs to know where the centromere and telomere regions start and end.
This data is accessible for many species from UCSC:

(image is an example, select relevant species/build)
Select the relevant species and build, along with the fields indicated above. You will need to set a filters, see following information also.
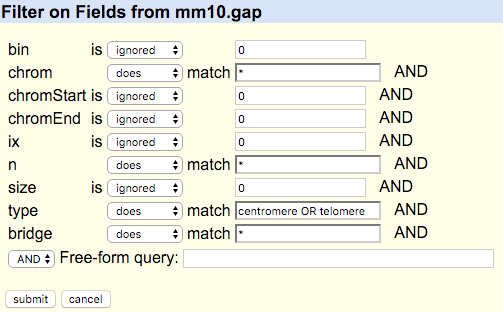
Unfortunately UCSC change the data in the tables quite regularly. In more recent builds it appears that there is a specific Centromeres track. If the above didn't give centromere outputs you will need to gather data from the new table getting min/max values for each chromosome.
Using the resulting data you construct a tab separated file following the format of this file:
chr ptel cen_start cen_end qtel comment
1 750000 121270001 150000000 249220001 .
2 10000 89330001 95390000 242950001 .
If you have no centromeres, set ptel and cen_start to 0, assigning the usable seq range to cen_end and qtel.
If you have no telomere or centromere data at all you can use this to generate the file:
export MITO='Mito'
perl -ane 'printf qq{%s\t0\t0\t1\t%d\t.\n},$F[0],$F[1];' genome.fa.fai | grep -v $MITO > centTelo.tsv