-
Notifications
You must be signed in to change notification settings - Fork 1
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
1 parent
555dc81
commit b092809
Showing
33 changed files
with
47,973 additions
and
133,881 deletions.
There are no files selected for viewing
This file was deleted.
Oops, something went wrong.
Large diffs are not rendered by default.
Oops, something went wrong.
Large diffs are not rendered by default.
Oops, something went wrong.
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,217 @@ | ||
| # Tips for spatiotemporal indexing | ||
|
|
||
|
|
||
| In the both the [AdaSTEM Demo](https://chenyangkang.github.io/stemflow/Examples/01.AdaSTEM_demo.html) and [SphereAdaSTEM demo](https://chenyangkang.github.io/stemflow/Examples/04.SphereAdaSTEM_demo.html) we use bird observation data to demonstrate functionality of AdaSTEM. Spatiotemporal coordinate are homogeneously encoded in these two cases, with `longitude` and `latitude` being spatial indexes and `DOY` (day of year) being temporal index. | ||
|
|
||
| Here, we present more tips and examples on how to play with these indexing systems. | ||
|
|
||
| ------ | ||
|
|
||
|
|
||
| ## 2D + Temporal indexing | ||
|
|
||
| ### Flexible coordinate systems | ||
|
|
||
| `stemflow` support all types of spatial coordinate reference system (CRS) and temporal indexing (for example, week month, year, or decades). `stemflow` only support tabular point data currently. You should transform your data to desired CRS before feeding them to `stemflow`. | ||
|
|
||
| For example, transforming CRS: | ||
|
|
||
| ```python | ||
| import pyproj | ||
|
|
||
| # Define the source and destination coordinate systems | ||
| source_crs = pyproj.CRS.from_epsg(4326) # WGS 84 (latitude, longitude) | ||
| target_crs = pyproj.CRS.from_string("ESRI:54017") # World Behrmann equal area projection (x, y) | ||
|
|
||
| # Create a transformer object | ||
| transformer = pyproj.Transformer.from_crs(source_crs, target_crs, always_xy=True) | ||
|
|
||
| # Project | ||
| data['proj_lng'], data['proj_lat'] = transformer.transform(data['lng'].values, data['lat'].values) | ||
| ``` | ||
|
|
||
| Now the projected spatial coordinate for each record is stored in `data['proj_lng']` and `data['proj_lat']` | ||
| We can then feed this data to `stemflow`: | ||
|
|
||
|
|
||
|
|
||
|
|
||
| ```python | ||
|
|
||
| from stemflow.model.AdaSTEM import AdaSTEMClassifier | ||
| from xgboost import XGBClassifier | ||
|
|
||
| model = AdaSTEMClassifier( | ||
| base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), | ||
| save_gridding_plot = True, | ||
| ensemble_fold=10, # data are modeled 10 times, each time with jitter and rotation in Quadtree algo | ||
| min_ensemble_required=7, # Only points covered by > 7 stixels will be predicted | ||
| grid_len_upper_threshold=1e5, # force splitting if the edge of grid exceeds 1e5 meters | ||
| grid_len_lower_threshold=1e3, # stop splitting if the edge of grid fall short 1e3 meters | ||
| temporal_start=1, # The next 4 params define the temporal sliding window | ||
| temporal_end=52, | ||
| temporal_step=2, | ||
| temporal_bin_interval=4, | ||
| points_lower_threshold=50, # Only stixels with more than 50 samples are trained | ||
| Spatio1='proj_lng', # Use the column 'proj_lng' and 'proj_lat' as spatial indexes | ||
| Spatio2='proj_lat', | ||
| Temporal1='Week', | ||
| use_temporal_to_train=True, # In each stixel, whether 'Week' should be a predictor | ||
| njobs=1 | ||
| ) | ||
| ``` | ||
|
|
||
| Here, we use temporal bin of 4 weeks and step of 2 weeks, starting from week 1 to week 52. For spatial indexing, we force the gird size to be `1km (1e3 m) ~ 10km (1e5 m)`. Since `ESRI 54017` is an equal area projection, the unit is meter. | ||
|
|
||
|
|
||
| Then we could fit the model: | ||
|
|
||
| ```py | ||
| ## fit | ||
| model = model.fit(data.drop('target', axis=1), data[['target']]) | ||
|
|
||
| ## predict | ||
| pred = model.predict(X_test) | ||
| pred = np.where(pred<0, 0, pred) | ||
| eval_metrics = AdaSTEM.eval_STEM_res('classification',y_test, pred_mean) | ||
| ``` | ||
|
|
||
| Note that the [Quadtree [1]](https://dl.acm.org/doi/abs/10.1145/356924.356930) algo is limited to 6 digits for efficiency. So transform your coordinate of it exceeds that threshold. For example, x=0.0000001 and y=0.0000012 will be problematic. Consider changing them to x=100 and y=1200. | ||
|
|
||
| ------ | ||
| ### Spatial-only modeling | ||
|
|
||
| By playing some tricks, you can also do a `spatial-only` modeling, without splitting the data into temporal blocks: | ||
|
|
||
| ```python | ||
| model = AdaSTEMClassifier( | ||
| base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), | ||
| save_gridding_plot = True, | ||
| ensemble_fold=10, | ||
| min_ensemble_required=7, | ||
| grid_len_upper_threshold=1e5, | ||
| grid_len_lower_threshold=1e3, | ||
| temporal_start=1, | ||
| temporal_end=52, | ||
| temporal_step=1000, # Setting step and interval largely outweigh | ||
| temporal_bin_interval=1000, # temporal scale of data | ||
| points_lower_threshold=50, | ||
| Spatio1='proj_lng', | ||
| Spatio2='proj_lat', | ||
| Temporal1='Week', | ||
| use_temporal_to_train=True, | ||
| njobs=1 | ||
| ) | ||
| ``` | ||
|
|
||
| Setting `temporal_step` and `temporal_bin_interval` largely outweigh the temporal scale (1000 compared with 52) of your data will render only `one` temporal window during splitting. Consequently, your model would become a spatial model. This could be beneficial if temporal heterogeneity is not of interest, or without enough data to investigate. | ||
|
|
||
| ------- | ||
|
|
||
| ### Fix the gird size of Quadtree algorithm | ||
|
|
||
| There are **two ways** to fix the grid size: | ||
|
|
||
| #### 1. By using some tricks we can fix the gird size/edge length of AdaSTEM model classes: | ||
|
|
||
| ```python | ||
| model = AdaSTEMClassifier( | ||
| base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), | ||
| save_gridding_plot = True, | ||
| ensemble_fold=10, | ||
| min_ensemble_required=7, | ||
| grid_len_upper_threshold=1000, | ||
| grid_len_lower_threshold=1000, | ||
| temporal_start=1, | ||
| temporal_end=52, | ||
| temporal_step=2, | ||
| temporal_bin_interval=4, | ||
| points_lower_threshold=0, | ||
| stixel_training_size_threshold=50, | ||
| Spatio1='proj_lng', | ||
| Spatio2='proj_lat', | ||
| Temporal1='Week', | ||
| use_temporal_to_train=True, | ||
| njobs=1 | ||
| ) | ||
| ``` | ||
|
|
||
| Quadtree will keep splitting until it hits an edge length lower than 1000 meters. Data volume won't hamper this process because the splitting threshold is set to 0 (`points_lower_threshold=0`). Stixels with sample volume less than 50 still won't be trained (`stixel_training_size_threshold=50`). However, we cannot guarantee the exact grid length. It should be somewhere between 500m and 1000m since each time Quadtree do a bifurcated splitting. | ||
|
|
||
| #### 2. Using `STEM` model classes | ||
|
|
||
| We also implemented `STEM` model classes for fixed gridding. Instead of adaptive splitting based on data abundance, `STEM` model classes split the space with fixed grid length: | ||
|
|
||
| ```python | ||
| from stemflow.model.STEM import STEM, STEMRegressor, STEMClassifier | ||
|
|
||
| model = STEMClassifier( | ||
| base_model=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0,n_jobs=1), | ||
| save_gridding_plot = True, | ||
| ensemble_fold=10, | ||
| min_ensemble_required=7, | ||
| grid_len=1000, | ||
| temporal_start=1, | ||
| temporal_end=52, | ||
| temporal_step=2, | ||
| temporal_bin_interval=4, | ||
| points_lower_threshold=0, | ||
| stixel_training_size_threshold=50, | ||
| Spatio1='proj_lng', | ||
| Spatio2='proj_lat', | ||
| Temporal1='Week', | ||
| use_temporal_to_train=True, | ||
| njobs=1 | ||
| ) | ||
| ``` | ||
|
|
||
| Here, `grid_len` parameter take place the original upper and lower threshold parameters. The main functionality is the same as `AdaSTEM` classes. | ||
|
|
||
| ---- | ||
| ## 3D spherical + Temporal indexing | ||
|
|
||
| Our earth is a sphere, and consequently there is no single solution to project the sphere to a 2D plane while maintaining the distance and area – all projection method as pros and cons. We also implemented spherical indexing to solve this issue. | ||
|
|
||
|
|
||
| ```python | ||
| from stemflow.model.SphereAdaSTEM import SphereAdaSTEMRegressor | ||
| from xgboost import XGBClassifier, XGBRegressor | ||
|
|
||
| model = SphereAdaSTEMRegressor( | ||
| base_model=Hurdle( | ||
| classifier=XGBClassifier(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1), | ||
| regressor=XGBRegressor(tree_method='hist',random_state=42, verbosity = 0, n_jobs=1) | ||
| ), # hurdel model for zero-inflated problem (e.g., count) | ||
| save_gridding_plot = True, | ||
| ensemble_fold=10, # data are modeled 10 times, each time with jitter and rotation in Quadtree algo | ||
| min_ensemble_required=7, # Only points covered by > 7 stixels will be predicted | ||
| grid_len_upper_threshold=2500, # force splitting if the grid length exceeds 2500 (km) | ||
| grid_len_lower_threshold=500, # stop splitting if the grid length fall short 500 (km) | ||
| temporal_start=1, # The next 4 params define the temporal sliding window | ||
| temporal_end=366, | ||
| temporal_step=25, # The window takes steps of 20 DOY (see AdaSTEM demo for details) | ||
| temporal_bin_interval=50, # Each window will contain data of 50 DOY | ||
| points_lower_threshold=50, # Only stixels with more than 50 samples are trained | ||
| Temporal1='DOY', | ||
| use_temporal_to_train=True, # In each stixel, whether 'DOY' should be a predictor | ||
| njobs=1 | ||
| ) | ||
| ``` | ||
|
|
||
| `SphereAdaSTEM` module has almost the same structure and functions as `AdaSTEM` and `STEM` modules. The only difference is that | ||
|
|
||
| 1. It mandatorily looks for "longitude" and "latitude" in the columns. | ||
| 1. It splits the data using [`Sphere QuadTree` [2]](https://ieeexplore.ieee.org/abstract/document/146380). | ||
| 1. It plots the grids using `plotly`. | ||
|
|
||
|
|
||
| See [SphereAdaSTEM demo](https://chenyangkang.github.io/stemflow/Examples/04.SphereAdaSTEM_demo.html) and [Interactive spherical gridding plot](https://chenyangkang.github.io/stemflow/assets/Sphere_gridding.html). | ||
|
|
||
| 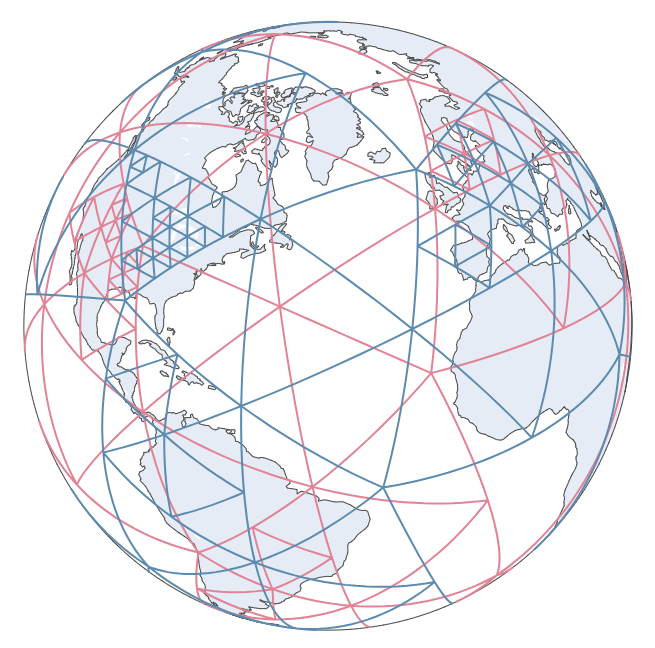 | ||
|
|
||
| ----- | ||
| ## References: | ||
|
|
||
| 1. [Samet, H. (1984). The quadtree and related hierarchical data structures. ACM Computing Surveys (CSUR), 16(2), 187-260.](https://dl.acm.org/doi/abs/10.1145/356924.356930) | ||
|
|
||
| 1. [Gyorgy, F. (1990, October). Rendering and managing spherical data with sphere quadtrees. In Proceedings of the First IEEE Conference on Visualization: Visualization90 (pp. 176-186). IEEE.](https://ieeexplore.ieee.org/abstract/document/146380) |
File renamed without changes
File renamed without changes
File renamed without changes.
File renamed without changes.
File renamed without changes
File renamed without changes.
File renamed without changes
File renamed without changes.
File renamed without changes
File renamed without changes
File renamed without changes
File renamed without changes
File renamed without changes
File renamed without changes
Oops, something went wrong.