Paper under review. Citation information at the end of the README
Note: Users of the processed benchmark datasets, which are the outputs generated by the code in this repository, must comply with the MIMIC-IV Data Use Agreement. Please refrain from distributing them to others. Instead, direct individuals to this repository and https://physionet.org/ for access. Never upload the benchmark dataset in publicly accessible locations, such as GitHub repositories.
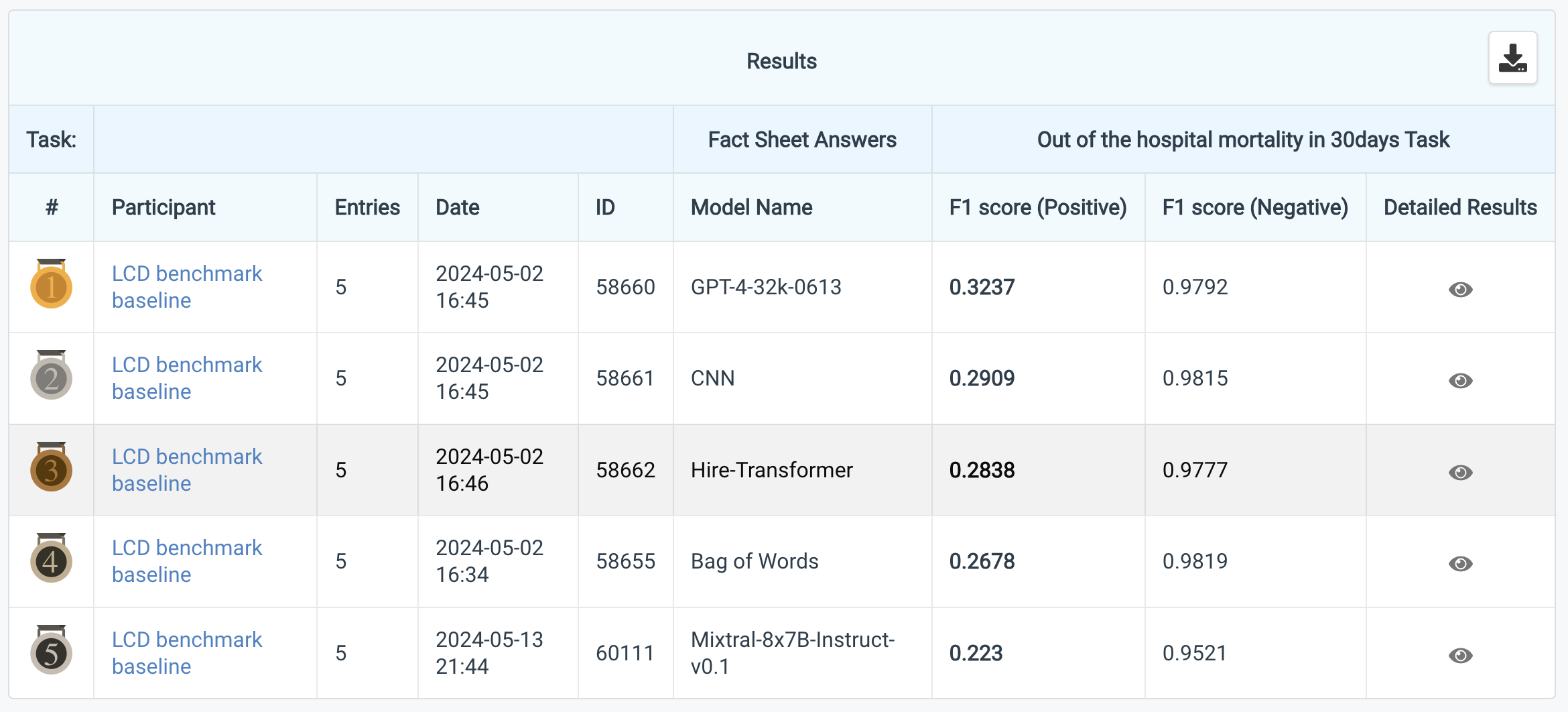
Link to LCD benchmark CodaBench
- LCD benchmark supports CodaBench, which is an online dataset evaluation platform. Please submit your model's output through CodaBench and compare it with others.
Metadata creation. (Pull req #5)
- Add code to generate metadata.json (with patient IDs for train/dev/test split) and update README with metadata details (See 6. of Steps)
Added subtasks for 60, 90 days. (Pull req #4)
- Added
--task_nameflag and modified README accordingly. - Changed the folder name for main task (30-day) from
out_hos_30days_mortality/labels.json→ toout_hospital_mortality_30/labels.json
LLM experiment example codes. (Pull req #1)
- For LLM experiment example codes, please see llm_experiment_examples folder
- cf. Codes for other baseline models (CNN and Hierarchical Transformer) are available on CNLPT library as stated in the paper.
Future plans:
- We are planning to expand our data distribution platforms (Full data and preprocessing code will soon be available on PhysioNet)
This repository only provides hashed (using one-way encryption) document ids and the labels of each datapoint. You will need to download notes from PhysioNet.org after fully executed MIMIC-IV data use agreement. Agreeing to PhysioNet Credentialed Health Data Use Agreement 1.5.0 and PhysioNet Credentialed Health Data License 1.5.0 is required to utilize the benchmark dataset.
Please get access to both MIMIC-IV v2.2 and MIMIC-IV-Note v2.2.
We strongly recommend to use Python version 3.9 or higher.
The example codes are tested on Ubuntu 22.04 (python v3.10.12) and MacOS v13.5.2 (python v3.9.6).
pandas and tqdm are required. To install these: pip install -r requirements.txt
Please pay attention to the message (stdout) at the end of processing run, as it will tell the integrity of the created data. Note that the integrity does not check the order of the instances in datasets.
create_data.py : Code that merges labels.json with MIMIC-IV note data.
create_data_serverside.py : (Internal use only)Please note that create_data_serverside.py is the code the authors used to extract labels from pre-processed data. It is not required for users to run the code.
-
Complete the steps required by MIMIC-IV-Note v2.2 and download
mimic-iv-note-deidentified-free-text-clinical-notes-2.2.zipfile. -
Unzip the downloaded file. Following is an example bash script for linux users :
#cd <MOVE_TO_DOWNLOAD_FOLDER>
unzip mimic-iv-note-deidentified-free-text-clinical-notes-2.2.zip
cd note/
gzip -d discharge.csv.gz
export NOTE_PATH=${PWD}/discharge.csvThe path to discharge.csv (3.3G) is stored in $NOTE_PATH
shasum -a 1 discharge.csv # sha1 value
# -> c4f0cfcd00bb8cbb118b1613a5c93f31a361e82b discharge.csv
# Or a9ac402818385f6ab5a574b4516abffde95d641a discharge.csv (old version)- Clone this repository
# Move to any project folder
# e.g.) cd <MOVE_TO_ANY_PROJECT_FOLDER>
git clone https://github.com/Machine-Learning-for-Medical-Language/long-clinical-doc.git
cd long-clinical-doc- Merge notes and the labels
export TASK_NAME="out_hospital_mortality_30"
export LABEL_PATH=${TASK_NAME}/labels.json
export OUTPUT_PATH=${TASK_NAME}
python create_data.py \
--label_path ${LABEL_PATH} \
--discharge_path ${NOTE_PATH} \
--output_path ${OUTPUT_PATH} \
--task_name ${TASK_NAME}For the subtasks of 60-day or 90-day mortality, please change the TASK_NAME so that both the path and the --task_name argument are updated accordingly.
- Please make sure to check the number of processed datapoints. If the numbers match the values written below, then the contents of the datasets are identical to our version. (Note that this integrity check does not verify the order of instances in the datasets.)
Train: 34,759
Dev: 7,505
Test: 7,568
- Once processing is completed, metadata — including data_type (indicating whether the instance is train, dev, or test), note_id, subject_id, hadm_id, note_type, note_seq, charttime, and storetime — will be stored in
${OUTPUT_PATH}/${TASK_NAME}/metadata.json.
Example (numbers are replaced with arbitrary values):
{
"0xbfe5e112c39b6240f54dc3af123456": {
"note_id": "10000000-DS-01",
"subject_id": 10000000,
"hadm_id": 12345678,
"note_type": "DS",
"note_seq": 01,
"charttime": "2199-12-29 00:00:00",
"storetime": "2199-12-30 21:59:00",
"data_type": "train"
},
"0xd8e888974794dffcc8d72734567890": {
Citation information:
@article{yoon2024lcd,
title={LCD Benchmark: Long Clinical Document Benchmark on Mortality Prediction for Language Models},
author={Yoon, WonJin and Chen, Shan and Gao, Yanjun and Zhao, Zhanzhan and Dligach, Dmitriy and Bitterman, Danielle S and Afshar, Majid and Miller, Timothy},
journal={Under review (Preprint: medRxiv)},
pages={2024--03},
year={2024}
}